In 2017, I got a call from Ginger Hultin, my new health data coach. She was concerned, she said, about my TMAOs.
“My what?” I asked.
“Your TMAOs,” she repeated, referring to trimethylamine-N-oxide, a metabolite excreted by bacteria in the stomach that can increase risk for heart disease if levels get too high.
Who knew?
Not to worry, said Hultin in a soothing, upbeat voice. I could reduce my score by cutting back on red meat, which TMAO-secreting bacteria love to gorge on.
Trimethylamine-N-oxides were part of a battery of tests I had taken a few weeks earlier when Hultin’s employer, a Seattle start-up called Arivale, had collected copious amounts of my blood, saliva, and stool to test hundreds of biomarkers. These included DNA markers, proteins, metabolites, lipids like cholesterol, and the microbiome in my gut.
The company had also sent me a Fitbit to measure steps, sleep and heart rate. Online they had asked endless questions about my health, medical history, happiness, stress and more, to add to my digital health report card—information that was integrated with my other data using advanced computers and algorithms to produce the report.
The goal was for me, a basically hale and hearty man in my fifties, to find out just how healthy I really was—and would be.
Hultin asked me to scroll to a section called “Genes” in my online Arivale profile. “Do you see the finding about vitamin D?” she asked. My result for a gene called VDR indicated that I had a mutation that makes it difficult for my body to absorb vitamin D. “This is probably why your vitamin D level is low,” she said, referring to yet another section of my profile. Not dangerously so, though she suggested that I start taking a vitamin supplement.
I was impressed. I had spent years as a reporter trying out hundreds of newfangled tests like these to see what they might reveal about the health of an actual human, findings that I had chronicled in my 2009 book Experimental Man and in dozens of articles before and after, including a 2017 story in NEO.LIFE, “The Radical Idea of Avoiding Sickness”. Most of them, however, had been too new, experimental, and incomplete to tell me much.
Arivale’s data and analysis was different. It seemed more scientifically sound. More important, it seemed believable.
Yes, the company was testing just a small number of biodata points, a few hundred out of the thousands that might be influencing, say, my risk for heart disease. Nor was TMAO likely to have an immediate influence—or much influence at all, compared with other risk factors—on whether my heart would keep happily beating, or would one day seize up. Yet the report was telling me things that few people learn from standard exams. I was also being given choices based on my own specific data about how to intervene in my own healthcare—for instance, to rein in the burgers and BBQ pulled pork sandwiches or face the consequences.
At the time, I remember feeling like I had just gotten a checkup from the future, something that scientists and entrepreneurs had repeatedly promised me during my experimental man project, but seldom delivered on. This wasn’t surprising given the complexity of human biology and the newness of the science, although I had been wondering when all of this would finally come together to make a difference in keeping me in tip-top health.
Could the advent of Arivale be the moment?
As it turned out, it was not. Two years later, Arivale folded, a victim mostly of high costs. Few customers were willing to pay $3,400 for a profile, or even $99 a month, to which the company eventually slashed its price. But Arivale did signal the dawn, or perhaps the predawn, of a new era in health care that in 2022 seems even closer to being realized. It’s called scientific wellness, a field that collects and analyzes reams of biodata and uses it to keep you well rather than waiting for you to get sick before taking action.
One way to look at scientific wellness is to think of it as “well care,” rather than the “sick care” that is the current standard of care in the U.S. and the rest of the world. “Well care” means keeping a person healthy, which doesn’t at first glance seem radical. Yet it is, in part because the science to delve deeply into the secrets hidden in our genes and in other molecules like TMAO was, until recently, unavailable.
Now the data is arriving, even if much work remains to make sense of it. Scientific wellness may be poised at last to usher in a new health-care paradigm, in which people will routinely get Arivale-style tests and analysis. This will include getting a dashboard of risk factors like most people have never seen before that will track transitions from a state of wellness to a state of disease. It will be like getting weather forecasts about a personal future of possible maladies, and what can be done now to prevent them.
Scientists and physicians are already using scientific wellness approaches to help develop new and better diagnosis (or prediagnosis) methods and potentially improved treatments for everything from cancer to brain health.
“The science and technology to help us predict and prevent diseases is arriving,” says Leroy Hood, a physician and biologist who founded Arivale. Hood, 83, has been a leading figure in precision health for more than two decades, since long before the science was ready and the term “precision health” even existed. “The big task ahead of us now,” says Hood, “is to take all this and make it work for millions of people.”
A New Approach
To do this, Hood recently launched another endeavor to jump-start the age of scientific wellness that is much more ambitious than Arivale. It comes as other wellness efforts are gathering steam, including government-sponsored projects like biobanks that have been for years collecting DNA and other biodata on millions of people, and are just now starting to use new technologies and discoveries to make sense of it.
The private sector is also delving into treasure troves of new data, including drug companies that hope to use it to develop new pharmaceuticals. A handful of start-ups are also developing risk-profile tests and algorithms using biobank data. These include Genomics PLC, headquartered in Oxford, England, which is working with the Manchester-based, UK Biobank to develop risk scores for dozens of diseases, including heart disease, diabetes, and several cancers. The company is tapping into DNA, electronic medical records, and other biodata that was collected (with consent) from 500,000 Brits. Other fledgling businesses, such as Alden Scientific in Boston, are in the early stages of creating complex profiles of biodata for individual consumers.
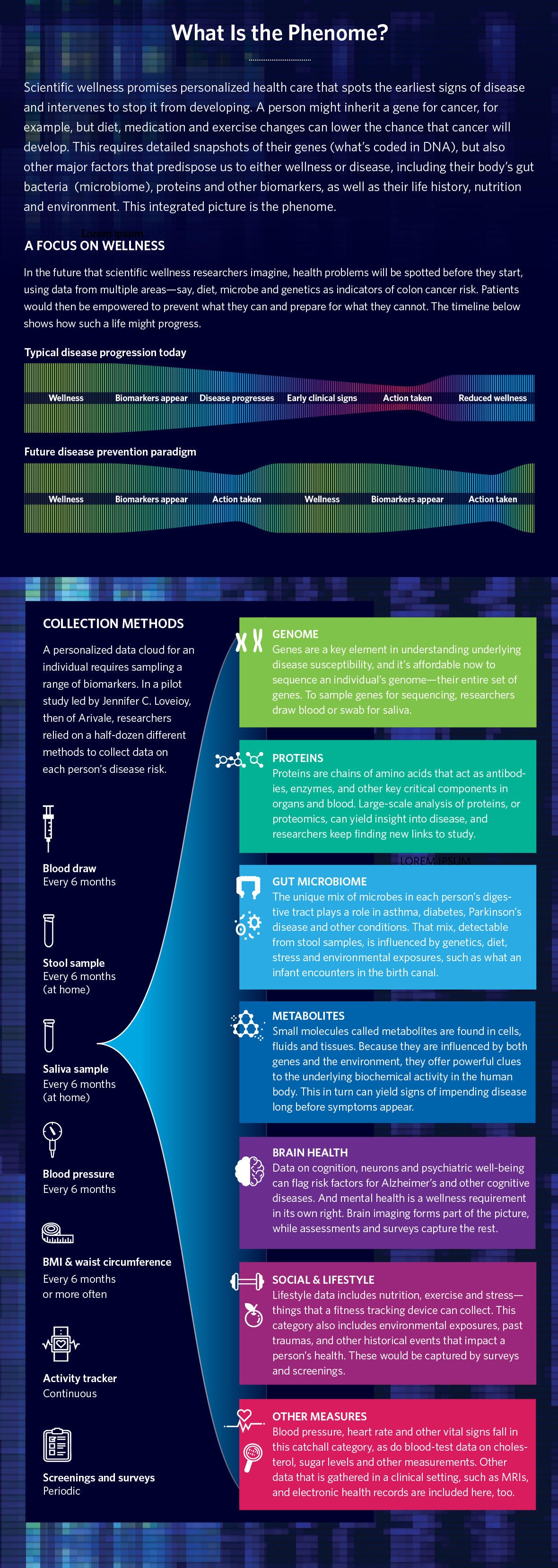
Hood’s new venture is called Phenome Health, a name derived from the term “phenomics”—the science of measuring one’s phenotype. This refers to a person’s state of health at any given moment as influenced by their genes and from changes in other molecules, such as proteins, in the body—plus the impact of diet, lifestyle, age, and other factors.
Phenome Health, says Hood, is a variation on Arivale, but with several key differences. “First of all, it’s a nonprofit. We realized that before all this will work commercially, there is still some basic science and research to be done, and economies of scale that need to happen to bring the costs down.” Hood also is planning to raise far more money than anyone else working in phenomics—a whopping $10 billion. He hopes to find it not from investors or even traditional grants but from the U.S. Congress—a big ask in Washington these days, but one Hood is convinced he can make happen.
Another lesson learned from Arivale is that they needed more people and more biomarkers to allow their analysis to draw firmer and more comprehensive conclusions about risk. “We need to include a lot more people,” says Hood, “and run them on a lot more tests,” which is why he wants to recruit one million volunteers to give samples in the U.S. for a project that will run for 10 years. His wish list of biodata includes not only sequences of complete genomes—a person’s entire DNA, which now costs around $600 per genome—but also thousands of proteins, those compounds in the body made by cells from instructions from DNA that include everything from enzymes and hormones to structural elements in skin, hair, fingernails and more. Phenome Health also plans to collect everything from bacteria in the gut to metabolites like TMAO, which are chemicals that our bodies—or bacteria living inside our bodies—create when they break down food, drugs and other chemicals.
“We’re planning to test for 3,000 proteins,” says Hood, far more than Arivale. “We’re probably going to do 2,500 metabolites,” he adds, also a deluge compared to past efforts, “and a whole variety of key clinical chemistries, including levels in your body of environmental toxins like mercury.” Phenome will collect data on brain health working with brain-testing company Posit Health, a partner that’s signed up to assess 25 cognitive functions, and to create a digital neural profile.
Recruits also will be asked to use wearables like Fitbits that measure not only steps, sleep and heart rates, but also oxygen levels, heart variability, erratic pulses like atrial fibrillations, and daily calories consumed. Plus, they will run standard blood tests like creatinine and cholesterol.
All this data will be crunched using advanced artificial intelligence and machine learning that also will incorporate subjects’ medical records, journal articles and other medical information, all to produce a state-of-the-art phenomics report card. Doctors will be able to use this report card to assess a person’s current and future health and to help their patients, as Ginger Hultin helped me using my Arivale data, to start any preventive measures that need to be taken.
A huge concern with collecting all this intimate health data is privacy. We live in an era when even banks and secure government data—not to mention health information collected by doctors, hospitals, insurers, and by health apps on our phones—are sometimes hacked. “Of course, we will institute state-of-the-art security measures to protect people’s data,” says Hood. Still, security breaches will remain a possibility, just as they do for people who do their banking on apps and conduct other aspects of their lives on digital systems.
A Long and Winding Road
Hood’s obsession with phenomics goes back more than 40 years to when he was at the California Institute of Technology in the 1970s and 1980s, when he first became interested in applying information technology to biology. This horrified his colleagues, who in those days preferred to work directly with organisms and ecosystems and didn’t yet realize how important computers and digital databases were about to become to their field. “My colleagues in the biology department tried to move me to engineering,” he remembers, even as he worked back then to co-invent some of the first advanced DNA sequencers. This included technology that later became the core to Applied Biosystems, the first major sequencing company, founded in 1981.
Parting ways with Caltech in 1991, Hood moved to Seattle, where he headed up the Department of Molecular Biotechnology at the University of Washington before founding the Institute for Systems Biology in 2000. This was another radical notion of his—that biologists should be studying entire systems of molecular and physiological activity in cells and organisms—and at a time when the norm was a reductive approach that isolated and studied single genetic biomarkers and highly focused functions and systems. Around this time Hood coined what he called the “Three P’s”—Personalized, Predictive, and Preventative—as an early way to describe his vision, and later added a fourth—Participatory—at the suggestion of Google co-founder Larry Page.
In the early 2000s, the science was driven mostly by genetics, as the Human Genome Project was finishing the first-ever sequence of a complete human genome, a 10-year-plus project funded by Congress for $2.7 billion ($5.5 billion in 2022 dollars). Excitement around DNA was at a fever pitch as geneticists discovered gene variants like APOE4, a mutation that is a strong indicator of Alzheimer’s disease, and BRCA1 and BRCA2, which indicate high risk for breast cancer. Biotech companies like Genentech were also developing new drugs such as Herceptin, which targets a genetic mutation in the HER gene that causes various cancers, including stomach, esophageal, and breast, shutting the gene down. Geneticists were also discovering genes for rare and often devastating diseases like cystic fibrosis and Fragile X syndrome.
Science in the early 2000s, however, remained a long way off from realizing Hood’s visions for scientific wellness. What was available using molecular markers was almost exclusively genomic and focused largely on single genetic letters—the A, T, C and G of DNA—and how differences in these letters, called single nucleotide polymorphisms (SNPs), seemed to be correlated with a higher risk for disease. For instance, a G instead of a C might slightly bump up a person’s risk of getting bladder cancer, although a few years later scientists discovered that the effect size of these single letter mutations in causing disease tended to be very small compared to other risk factors like age, diet, and other genes. Scientists back then were also talking about assessing the combined effects of multiple genes (polygenics) and biomarkers like proteins and metabolites. But using these multiple biomarkers for determining disease risk remained costly and mostly beyond that day’s technical capacity.
Only in the last decade did Arivale become scientifically feasible. Founded in 2015, it ultimately tested 5,000 people. Although this population wasn’t particularly large, given the ambitious goals, it was enough to get some preliminary results. “We were able to monitor 167 individuals as they transitioned from wellness to sickness over a four-year stretch,” he says. “We looked at ten of these people who then transitioned to cancer. In every case there were proteins that were way off the scale compared with the normal average. We mapped them into disease-perturbed networks and projected what the transition was going to be.”
Arivale proved the concept of scientific wellness, which is why Hood considers it to be a success even though it failed as a business.
Experiments to Come
Having proved the concept, the next task is to deepen the pool of data and expand the insights that can be gleaned from it. That will require building workable platforms, honing methods, gathering more baseline data and developing new and better technologies. Computers need to be taught how to recognize critical information within a firehose of unstructured data. Iya Khalil, a theoretical physicist turned bio-AI expert who is the global head of the AI Innovation Lab at Novartis, says that much more needs to be done. “This stuff is really complicated,” she says. “Right now, we’re better at using this data for rare diseases that are easier to identify and predict. We need to work on building it out for more common diseases that have multiple risk factors. But it’s becoming much more real than it was before.”
The current state of scientific wellness is analogous to the Human Genome Project in the 1990s, says Hood. Although it had a big price tag, it helped the nascent genomics industry scale up its capabilities, which ultimately brought costs down. “We think that will happen with phenomics,” he says.
“Prices are already falling,” says Rory Collins, professor of medicine and epidemiology at the University of Oxford and the chief executive of the UK Biobank, the government- and private philanthropy-sponsored project that has genetically sequenced 500,000 Britons, and is planning to add phenomics biomarkers to their scores that predict future risks for several diseases. This is part of the initiative developed by Genomics PLC, in Oxford, that recently announced polygenic risk profiles for 28 diseases. By using UK Biobank data, for instance, Genomics PLC was able to provide risk assessments for apparently healthy people—such as a 40-year-old man with no symptoms or family history of heart problems who, it was revealed, is at high risk for heart disease by the time he turns 60. Other biobanks, such as the Massachusetts General Brigham biobank at Harvard, are also developing polygenic risk profiles and plan to soon include proteins and other markers.
Phenome Health’s effort would be larger than any of these—if Hood succeeds in procuring twice the amount of funding that the Human Genome Project raised in the early 1990s. That’s a big if in an era when pushing through big science projects is politicized and fraught. But Hood is confident. Phenome is in discussions with “key members of Congress,” he says. It is also exploring partnerships with companies and organizations such as Guardian Research Network, which has a patient base of 30 million patients in 13 states.
“I like the nonprofit approach Lee is taking,” says Khalil. He’s looking to power a new ecosystem, and not looking for ‘How do I make money off this data?’” Without such a basic effort, she adds, “we will continue doing health care in the same way we do it now.”
Hood suspects there will be a consumer-based “phenomics-and-me” style company spinning out from Phenome Health in future years when costs come down and more data is collected and analyzed. Right now, Phenome is working on a spin-out company based on the modular computer platform they’re building to analyze the data they’re planning to collect.
Some scientists ask why Phenome Health needs to recruit one million new people to study when dozens of biobanks and institutions around the world already have recruited and genetically sequenced millions of people. UK Biobank has now finished complete sequences of their entire cohort of 500,000 people. “There is a risk that setting up a new cohort will mean waiting another 10 to 15 years to get data that we already have tracked for over a decade,” says Collins. “It might make more sense to look at these cohorts that have already existed for a long time, and then think about which ones you might enhance with more depth in order to create your million or your two million.”
Hood counters that a new cohort can set out from the beginning to use the latest technology and build into it continual updates as they happen. Also, most risk profiles coming out of biobanks have tests for perhaps a few hundred thousand or a million SNPs— those single letter genetic markers—instead of nearly every one of the six billion nucleotides that come with sequencing a complete human genome, which is what Hood plans to do. “I think most of the SNP data is utterly trivial and it’s totally inadequate for what you can do now. No one is collecting as much data as we plan to,” he says.
However, the UK Biobank’s Collins confirmed that they have now finished complete sequences of their entire cohort of 500,000 people. (Data from the first 150,000 was released earlier this year in a study in Nature, which announced the discovery of almost a half-billion new genetic variants, far more than were known before.)
Diversity is another challenge for many biobanks. Most existing repositories include data from overwhelmingly white populations, which misses the rich diversity of human genetics. To remedy this shortfall, Phenome Health plans to work with its partner, Guardian, to tap into their large cohort of Black, Hispanic, and other marginalized people in the U.S. It’s also a social justice issue, given that different ethnicities have different genetic proclivities—variations that need to be better understood to improve prediction and treatment. “We need greater diversity to better understand the human gene pool,” Hood says.
Another hurdle to bringing scientific wellness to more people is a resistance to deploying phenomics in the clinic. “There’s a lag between the development of technologies and the ability to convince doctors and health-care systems that those technologies are valid and worthwhile, and sufficiently cost-effective or valuable to incorporate them into health care,” says Robert Green, a medical geneticist at Mass General Brigham Hospital, and a professor of medicine at Harvard Medical School, who studies how to bridge the gap between novel technologies and clinical practice. For instance, women are not routinely tested for BRCA1 and 2 genes to determine their breast cancer risk. These tests are available and well-established, but they have yet to be integrated into the daily workflow of most physicians. “How do you fit this into the 15 minutes that doctors have with their patients?” asks Green.
Standards are also lacking on how much evidence is needed to prove that a predictive test is good enough to be used routinely in the clinic. Nor are set pathways and rules established for how the FDA evaluates predictive DNA and other molecular tests. “We’ve got to get there,” says Green, “but the challenge is implementation, not vision.”
The way to gain acceptance, Hood believes, is to convince a few key physicians to sign on. That starts with working with them in demonstration projects and clinical trials. But he isn’t looking just to doctors and other providers to drive the shift to well care. He also expects patients to drive change, too. “Once well care reaches a critical mass of new people who taste its benefits,” he says, “they are going to see how transformational this type of health care is, and they’re going to demand it.”
With Phenome Health, Hood is convinced that his long quest to bring precision health to millions of people may be finally on the brink. “I’m absolutely convinced of it,” he says, with his trademark broad smile, intensity and optimism.
If he’s right, it may not be too long until everyone will have a Ginger Hultin calling them to discuss their TMAO and genetically influenced vitamin D levels, or whatever bubbles up from their own report. After I got Hultin’s news about my TMAO, I opted to cut back on the burgers and BBQ pulled pork sandwiches. A few months later, she called back, her voice cheery as ever, to go over my results from a fresh round of testing. My TMAO score, she reported, had shifted to normal.
With her help, I managed to make at least a small gesture towards staving off heart disease, now and, hopefully, in the future.
David Ewing Duncan is a journalist who writes for Vanity Fair, Wired, MIT Technology Review, the New York Times, the Atlantic and other publications. He is the author of ten books, most recently, Talking to Robots: Tales from Our Human-Robot Futures (Dutton).
Find out more about Phenome Health’s efforts to transform the future of health care here. Learn more about phenomics, the new science of wellness, in other stories in this special report.



